SBML Import: MAPK Signaling
Persistent Identifier
Use this permanent link to cite or share this Morpheus model:
Introduction
This model has been imported and automatically converted from SBML format by Morpheus. It shows oscillations in the MAPK signaling cascade (Kholodenko, 2000).
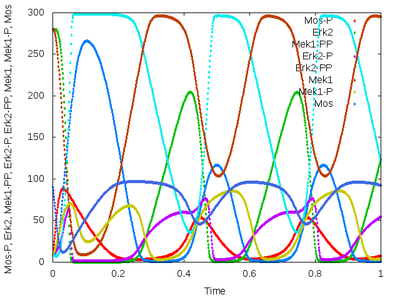
Description
Upon importing an SBML file, a Morpheus model is automatically created. A System of DiffEqns is generated, based on the function and reactions defined in the SBML file and defined as part of a CellType. Additionally, a Logger is generated to record and visualize the output.
Simulation details, such as StartTime and StopTime, as well as the time-step of System, need to be specified manually.
Things to try
- Browse the Biomodels database and try importing some SBML models.
Reference
B. N. Kholodenko: Negative feedback and ultrasensitivity can bring about oscillations in the mitogen-activated protein kinase cascades. Eur. J. Biochem. 267 (6): 1583-1588, 2000.
Model
Examples → ODE → MAPK_SBML.xml or
MAPK_SBML.xml
XML Preview
<?xml version='1.0' encoding='UTF-8'?>
<MorpheusModel version="3">
<Description>
<Title>BIOMD0000000010</Title>
<Details>Imported SBML model: http://www.ebi.ac.uk/biomodels-main/BIOMD0000000010
Kholodenko BN, Negative feedback and ultrasensitivity can bring about oscillations in the mitogen-activated protein kinase cascades.</Details>
</Description>
<Global/>
<Space>
<Lattice class="linear">
<Size symbol="size" value="1 0 0"/>
<Neighborhood>
<Order>1</Order>
</Neighborhood>
</Lattice>
<SpaceSymbol symbol="space"/>
</Space>
<Time>
<StartTime value="0"/>
<StopTime symbol="stop" value="1.0"/>
<TimeSymbol symbol="time" name="time"/>
</Time>
<CellTypes>
<CellType class="biological" name="sbml_ct">
<System solver="runge-kutta-adaptive" time-scaling="4000" time-step="stop">
<Constant symbol="V1" value="2.5"/>
<Constant symbol="Ki" value="9"/>
<Constant symbol="n" value="1"/>
<Constant symbol="K1" value="10"/>
<DiffEqn symbol-ref="MKKK" name="gained from reactions">
<Expression> - V1 * MKKK / ((1 + pow(MAPK_PP / Ki, n)) * (K1 + MKKK)) + V2 * MKKK_P / (KK2 + MKKK_P)</Expression>
</DiffEqn>
<DiffEqn symbol-ref="MKKK_P" name="gained from reactions">
<Expression>V1 * MKKK / ((1 + pow(MAPK_PP / Ki, n)) * (K1 + MKKK)) - V2 * MKKK_P / (KK2 + MKKK_P)</Expression>
</DiffEqn>
<Constant symbol="V2" value="0.25"/>
<Constant symbol="KK2" value="8"/>
<Constant symbol="k3" value="0.025"/>
<Constant symbol="KK3" value="15"/>
<DiffEqn symbol-ref="MKK" name="gained from reactions">
<Expression> - k3 * MKKK_P * MKK / (KK3 + MKK) + V6 * MKK_P / (KK6 + MKK_P)</Expression>
</DiffEqn>
<DiffEqn symbol-ref="MKK_P" name="gained from reactions">
<Expression>k3 * MKKK_P * MKK / (KK3 + MKK) - uVol * k4 * MKKK_P * MKK_P / (KK4 + MKK_P) + uVol * V5 * MKK_PP / (KK5 + MKK_PP) - V6 * MKK_P / (KK6 + MKK_P)</Expression>
</DiffEqn>
<Constant symbol="k4" value="0.025"/>
<Constant symbol="KK4" value="15"/>
<DiffEqn symbol-ref="MKK_PP" name="gained from reactions">
<Expression>k4 * MKKK_P * MKK_P / (KK4 + MKK_P) - V5 * MKK_PP / (KK5 + MKK_PP)</Expression>
</DiffEqn>
<Constant symbol="V5" value="0.75"/>
<Constant symbol="KK5" value="15"/>
<Constant symbol="V6" value="0.75"/>
<Constant symbol="KK6" value="15"/>
<Constant symbol="k7" value="0.025"/>
<Constant symbol="KK7" value="15"/>
<DiffEqn symbol-ref="MAPK" name="gained from reactions">
<Expression> - k7 * MKK_PP * MAPK / (KK7 + MAPK) + V10 * MAPK_P / (KK10 + MAPK_P)</Expression>
</DiffEqn>
<DiffEqn symbol-ref="MAPK_P" name="gained from reactions">
<Expression> k7 * MKK_PP * MAPK / (KK7 + MAPK) - k8 * MKK_PP * MAPK_P / (KK8 + MAPK_P) + uVol * V9 * MAPK_PP / (KK9 + MAPK_PP) - V10 * MAPK_P / (KK10 + MAPK_P)</Expression>
</DiffEqn>
<Constant symbol="k8" value="0.025"/>
<Constant symbol="KK8" value="15"/>
<DiffEqn symbol-ref="MAPK_PP" name="gained from reactions">
<Expression>k8 * MKK_PP * MAPK_P / (KK8 + MAPK_P) - V9 * MAPK_PP / (KK9 + MAPK_PP)</Expression>
</DiffEqn>
<Constant symbol="V9" value="0.5"/>
<Constant symbol="KK9" value="15"/>
<Constant symbol="V10" value="0.5"/>
<Constant symbol="KK10" value="15"/>
</System>
<Constant symbol="uVol" value="1" name="compartment size"/>
<Property symbol="MKKK" value="90" name="Mos"/>
<Property symbol="MKKK_P" value="10" name="Mos-P"/>
<Property symbol="MKK" value="280" name="Mek1"/>
<Property symbol="MKK_P" value="10" name="Mek1-P"/>
<Property symbol="MKK_PP" value="10" name="Mek1-PP"/>
<Property symbol="MAPK" value="280" name="Erk2"/>
<Property symbol="MAPK_P" value="10" name="Erk2-P"/>
<Property symbol="MAPK_PP" value="10" name="Erk2-PP"/>
</CellType>
</CellTypes>
<CellPopulations>
<Population size="1" type="sbml_ct"/>
</CellPopulations>
<Analysis>
<Logger time-step="stop/200">
<Restriction>
<Celltype celltype="sbml_ct"/>
</Restriction>
<Input>
<Symbol symbol-ref="MAPK"/>
<Symbol symbol-ref="MAPK_P"/>
<Symbol symbol-ref="MAPK_PP"/>
<Symbol symbol-ref="MKK"/>
<Symbol symbol-ref="MKK_P"/>
<Symbol symbol-ref="MKK_PP"/>
<Symbol symbol-ref="MKKK"/>
<Symbol symbol-ref="MKKK_P"/>
</Input>
<Output>
<TextOutput/>
</Output>
<Plots>
<Plot time-step="-1">
<Style grid="true" style="lines" line-width="3.0"/>
<Terminal terminal="png"/>
<X-axis>
<Symbol symbol-ref="time"/>
</X-axis>
<Y-axis>
<Symbol symbol-ref="MAPK"/>
<Symbol symbol-ref="MAPK_P"/>
<Symbol symbol-ref="MAPK_PP"/>
<Symbol symbol-ref="MKK"/>
<Symbol symbol-ref="MKK_P"/>
<Symbol symbol-ref="MKK_PP"/>
<Symbol symbol-ref="MKKK"/>
<Symbol symbol-ref="MKKK_P"/>
</Y-axis>
</Plot>
</Plots>
</Logger>
</Analysis>
</MorpheusModel>
Downloads
Files associated with this model: