Stem Cells in the Intestinal Crypt
Demonstrates the Morpheus features asymmetric cell division (
CellDivision), conditional change of cell type (ChangeCellType) and loading a simulation domain from anImage.
crypt.tiff.
Introduction
This illustrative example shows the emergence of clonal populations from stem cells in an intestinal crypt. Stem cells in the bottom of the crypt divide asymmetrically and produce a population of transit amplifying (TA) cells. For each TA cell, the color indicates the stem cell from which it was derived.
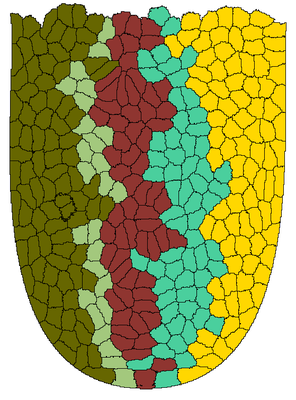
Description
The model shows several new modeling features, available as of Morpheus 1.2.
Loading Domain from Image
The crypt-like domain is specified by loading an external 8-bit TIFF image file crypt.tiff using the Image feature.
Asymmetric Cell Division
Stem cells divide asymmetrically using the new daughterID handles in the CellDivision element. This sets a user-defined symbol (here called daughter) to either daughter to
Conditionally Changing Cell Types
When a cell looses its stemness ChangeCellType element.
Upon satisfying its Condition, ChangeCellType moves the cell to the specified new cell type. By default, all the properties of a cell that exist in both cell type contexts are maintained, and unspecified ones are set to their default values. This default behavior can be overridden using Triggers that specify Rules stating how to deal with specific properties.
Mapper
Mappers allow the collection of statistical data about the cell population. Here, they are used to count the sizes of the various clonal populations. This number is reported into global Variables and subsequently written to a file and plotted using a Logger.
Model
Examples → CPM → Crypt.xml or
Crypt.xml
XML Preview
<?xml version='1.0' encoding='UTF-8'?>
<MorpheusModel version="4">
<Description>
<Title>Example-Crypt</Title>
<Details>Model ID: https://identifiers.org/morpheus/M0026
Full title: Stem Cells in the Intestinal Crypt
Date: 01.03.2016
Software: Morpheus (open source). Download from: https://morpheus.gitlab.io
Comment: Shows the following features of Morpheus: asymmetric cell division (proliferation), conditionally change cell type (ChangeCellType), loading simulation domain from image (Lattice/Domain).</Details>
</Description>
<Global>
<Variable symbol="w_d" name="wait time division" value="3000"/>
<Variable symbol="num_A" name="Clone A" value="0.0"/>
<Variable symbol="num_B" name="Clone B" value="0.0"/>
<Variable symbol="num_C" name="Clone C" value="0.0"/>
<Variable symbol="num_D" name="Clone D" value="0.0"/>
<Variable symbol="num_E" name="Clone E" value="0.0"/>
<Constant symbol="s" value="0.0"/>
<Constant symbol="clone" value="0.0"/>
</Global>
<Space>
<Lattice class="square">
<Size symbol="size" value="600 600 0"/>
<BoundaryConditions>
<Condition type="periodic" boundary="x"/>
<Condition type="periodic" boundary="y"/>
</BoundaryConditions>
<Neighborhood>
<Order>2</Order>
</Neighborhood>
<Domain boundary-type="noflux">
<Image path="crypt.tiff"/>
</Domain>
</Lattice>
<SpaceSymbol symbol="l"/>
</Space>
<Time>
<StartTime value="0"/>
<StopTime value="50000"/>
<TimeSymbol symbol="time"/>
</Time>
<CellTypes>
<CellType class="biological" name="stem_cells">
<Property symbol="clone" value="0.0"/>
<Property symbol="t_d" name="time of division" value="0.0"/>
<Property symbol="s" name="stemness" value="1"/>
<VolumeConstraint target="800" strength="1"/>
<SurfaceConstraint target="1" strength="1" mode="aspherity"/>
<ChangeCellType newCellType="TA_cells">
<Condition>s==0</Condition>
<Triggers/>
</ChangeCellType>
<DirectedMotion direction="0, -1, 0" strength="0.5"/>
<CellDivision division-plane="random" daughterID="daughter">
<Condition>time > t_d</Condition>
<Triggers>
<Rule symbol-ref="s">
<Expression>if( daughter == 1, 1, 0 )</Expression>
</Rule>
<Rule symbol-ref="t_d">
<Expression>time + rand_norm(w_d,200)</Expression>
</Rule>
</Triggers>
</CellDivision>
</CellType>
<CellType class="biological" name="TA_cells">
<Property symbol="clone" value="0.0"/>
<Property symbol="t_d" name="time of division" value="0"/>
<Property symbol="d" name="divisions" value="0"/>
<VolumeConstraint target="600 " strength="1"/>
<SurfaceConstraint target="0.9" strength="1" mode="aspherity"/>
<CellDivision division-plane="random" daughterID="daughter">
<Condition>time > t_d</Condition>
<Triggers>
<Rule symbol-ref="d">
<Expression>d+0.5</Expression>
</Rule>
<Rule symbol-ref="t_d">
<Expression>time + rand_norm(w_d,500)</Expression>
</Rule>
</Triggers>
</CellDivision>
<CellDeath>
<Condition>if( cell.center.y > size.y - 20, 1, 0)</Condition>
</CellDeath>
<Mapper name="Count Clone A">
<Input value="clone==1"/>
<Output symbol-ref="num_A" mapping="sum"/>
</Mapper>
<Mapper name="Count Clone B">
<Input value=" clone==2"/>
<Output symbol-ref="num_B" mapping="sum"/>
</Mapper>
<Mapper name="Count Clone C">
<Input value="clone==3"/>
<Output symbol-ref="num_C" mapping="sum"/>
</Mapper>
<Mapper name="Count Clone D">
<Input value="clone==4"/>
<Output symbol-ref="num_D" mapping="sum"/>
</Mapper>
<Mapper name="Count Clone E">
<Input value="clone==5"/>
<Output symbol-ref="num_E" mapping="sum"/>
</Mapper>
</CellType>
</CellTypes>
<CPM>
<Interaction default="0">
<Contact type1="stem_cells" type2="stem_cells" value="-10"/>
<Contact type1="stem_cells" type2="TA_cells" value="10"/>
</Interaction>
<MonteCarloSampler stepper="edgelist">
<MCSDuration value="1.0"/>
<Neighborhood>
<Order>2</Order>
</Neighborhood>
<MetropolisKinetics yield="0.1" temperature="3"/>
</MonteCarloSampler>
<ShapeSurface scaling="norm">
<Neighborhood>
<Distance>2.5</Distance>
</Neighborhood>
</ShapeSurface>
</CPM>
<CellPopulations>
<Population type="stem_cells" size="0">
<InitRectangle number-of-cells="5" mode="regular">
<Dimensions origin="250.0, 75.0, 0.0" size="100,30,0"/>
</InitRectangle>
<InitProperty symbol-ref="clone">
<Expression>cell.id</Expression>
</InitProperty>
<InitProperty symbol-ref="t_d">
<Expression>rand_uni(0,w_d)</Expression>
</InitProperty>
</Population>
<Population type="TA_cells" size="0">
<InitRectangle number-of-cells="500" mode="regular">
<Dimensions origin="0,80, 0" size="600, 490, 0"/>
</InitRectangle>
<InitProperty symbol-ref="t_d">
<Expression>rand_uni(0,w_d)</Expression>
</InitProperty>
</Population>
</CellPopulations>
<Analysis>
<Gnuplotter time-step="250" decorate="false">
<Terminal name="png"/>
<Plot>
<Cells value="clone">
<ColorMap>
<Color value="20" color="gold"/>
<Color value="19" color="dark-pink"/>
<Color value="18" color="dark-khaki"/>
<Color value="17" color="dark-goldenrod"/>
<Color value="16" color="cyan"/>
<Color value="15" color="coral"/>
<Color value="14" color="chartreuse"/>
<Color value="13" color="brown4"/>
<Color value="12" color="bisque"/>
<Color value="11" color="beige"/>
<Color value="10" color="light-red"/>
<Color value="9" color="light-green"/>
<Color value="8" color="light-blue"/>
<Color value="7" color="gray"/>
<Color value="6" color="black"/>
<Color value="5" color="yellow"/>
<Color value="4" color="blue"/>
<Color value="3" color="green"/>
<Color value="2" color="red"/>
</ColorMap>
</Cells>
</Plot>
</Gnuplotter>
<Logger time-step="100">
<Input>
<Symbol symbol-ref="num_A"/>
<Symbol symbol-ref="num_B"/>
<Symbol symbol-ref="num_C"/>
<Symbol symbol-ref="num_D"/>
<Symbol symbol-ref="num_E"/>
</Input>
<Output>
<TextOutput/>
</Output>
<Plots>
<Plot time-step="5000" title="Clone numbers">
<Style grid="true" point-size="1" line-width="3.0" style="linespoints"/>
<Terminal terminal="png"/>
<X-axis>
<Symbol symbol-ref="time"/>
</X-axis>
<Y-axis>
<Symbol symbol-ref="num_A"/>
<Symbol symbol-ref="num_B"/>
<Symbol symbol-ref="num_C"/>
<Symbol symbol-ref="num_D"/>
<Symbol symbol-ref="num_E"/>
</Y-axis>
</Plot>
</Plots>
</Logger>
<ModelGraph format="svg" reduced="false" include-tags="#untagged"/>
</Analysis>
</MorpheusModel>
crypt.tiff.

Downloads
Files associated with this model: