Coupling CPM and PDE: Vascular Patterning
Introduction
This example shows a model of vascular network formation by paracrine signaling (Köhn-Luque et al., 2011) and employs a coupled CPM and reaction-diffusion model.
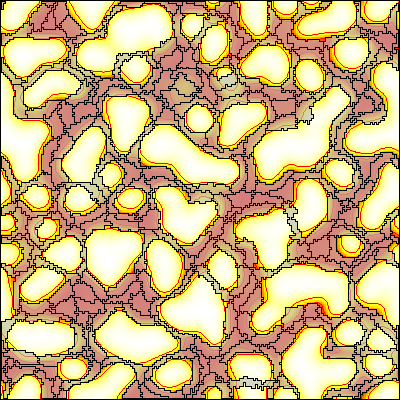
Description
The model defines a CPM as well as a PDE. These models are coupled by two processes:
- Cells, specified in
CellTypes, respond chemotactically to aLayer(or species) in thePDE. - Conversely, the production term of one
PDE Layeris coupled to the presence/absence of cell.
Reference
A. Köhn-Luque, W. de Back, J. Starruß, A. Mattiotti, A. Deutsch, J. M. Pérez-Pomares: Early Embryonic Vascular Patterning by Matrix-Mediated Paracrine Signalling: A Mathematical Model Study. PLoS ONE 6 (9): e24175, 2011.
Model
Get this model via:
Examples → Multiscale → VascularPatterning.xml or
VascularPatterning.xml
XML Preview
<MorpheusModel version="3">
<Description>
<Title>Example-VascularPatterning</Title>
<Details>Reference:
- A Köhn-Luque, W de Back, Y Yamaguchi, K Yoshimura, M A Herrero, T Miura (2013) Dynamics of VEGF matrix-retention in vascular network patterning, Physical Biology, 10(6) : 066007
http://dx.doi.org/10.1088/1478-3975/10/6/066007
Related to:
- Köhn-Luque A, de Back W, Starruß J, Mattiotti A, Deutsch A, Perez-Pomares JM, Herrero MA (2011) Early Embryonic Vascular Patterning by Matrix-Mediated Paracrine Signalling: A Mathematical Model Study. PLoS ONE 6(9): e24175.
http://dx.doi.org/10.1371/journal.pone.0024175
</Details>
</Description>
<Global>
<Field symbol="u" value="1.5e-6" name="VEGF">
<Diffusion rate="58.7"/>
</Field>
<Field symbol="s" value="0" name="Free ECM">
<Diffusion rate="0.001"/>
</Field>
<Field symbol="b" value="0" name="VEGF_b">
<Diffusion rate="0"/>
</Field>
<Field symbol="VEGF_all" value="0" name="VEGF_s + VEGF_b">
<Diffusion rate="0"/>
</Field>
<System solver="heun" time-step="5.0">
<Constant symbol="gamma" value="5e-3" name="Production ECM"/>
<Constant symbol="k_on" value="8.5e-4" name="Binding rate VEGF/ECM"/>
<Constant symbol="k_off" value="3.6e-3" name="Unbinding rate VEGF/ECM"/>
<Constant symbol="delta" value="2.6e-6" name="Decay VEGF "/>
<DiffEqn symbol-ref="u">
<Expression>- k_on*u*s + k_off*b - delta*u</Expression>
</DiffEqn>
<DiffEqn symbol-ref="s">
<Expression>gamma*cell - k_on*u*s+k_off*b</Expression>
</DiffEqn>
<DiffEqn symbol-ref="b">
<Expression>k_on*u*s - k_off*b</Expression>
</DiffEqn>
<Rule symbol-ref="VEGF_all">
<Expression>u+b</Expression>
</Rule>
</System>
<Constant symbol="cell" value="0.0"/>
<Constant symbol="cell_density" value="0.0045"/>
</Global>
<Space>
<Lattice class="square">
<Size symbol="size" value="200 200 0"/>
<BoundaryConditions>
<Condition boundary="x" type="periodic"/>
<Condition boundary="y" type="periodic"/>
</BoundaryConditions>
<NodeLength value="2"/>
<Neighborhood>
<Order>2</Order>
</Neighborhood>
</Lattice>
<SpaceSymbol symbol="l"/>
</Space>
<Time>
<StartTime value="0"/>
<StopTime value="3600"/>
<SaveInterval value="0"/>
<RandomSeed value="1"/>
<TimeSymbol symbol="t"/>
</Time>
<CellTypes>
<CellType class="biological" name="HUVEC">
<Property symbol="cell" value="1.0" name="cell"/>
<Property symbol="str" value="3e7" name="chemotactic strength"/>
<VolumeConstraint target="90" strength="1"/>
<Chemotaxis field="b" contact-inhibition="false" strength="str" retraction="false"/>
<!-- <Disabled>
<AddCell mode="exclude">
<Condition>rand_uni(0,1) < 0.24 + 0.0*t</Condition>
<Triggers/>
<Distribution>l.x / size.x</Distribution>
</AddCell>
</Disabled>
-->
</CellType>
<CellType class="medium" name="medium"/>
</CellTypes>
<CPM>
<Interaction default="0">
<Contact type1="medium" type2="HUVEC" value="3.2"/>
<Contact type1="HUVEC" type2="HUVEC" value="6.4"/>
</Interaction>
<MonteCarloSampler stepper="edgelist">
<MCSDuration value="1.0"/>
<Neighborhood>
<Order>2</Order>
</Neighborhood>
<MetropolisKinetics temperature="1"/>
</MonteCarloSampler>
<ShapeSurface scaling="norm">
<Neighborhood>
<Order>2</Order>
</Neighborhood>
</ShapeSurface>
</CPM>
<CellPopulations>
<Population size="0" type="HUVEC">
<InitRectangle mode="regular" number-of-cells="cell_density * size.x * size.y">
<Dimensions size="200,200,0" origin="0,0,0"/>
</InitRectangle>
</Population>
</CellPopulations>
<Analysis>
<Gnuplotter time-step="250" decorate="false">
<Terminal opacity="0.65" name="png"/>
<Plot>
<Field symbol-ref="b"/>
<Cells value="cell">
<ColorMap>
<Color value="1" color="gray"/>
<Color value="0.0" color="gray"/>
</ColorMap>
</Cells>
</Plot>
</Gnuplotter>
<!-- <Disabled>
<HistogramLogger normalized="false" number-of-bins="10" time-step="100">
<Plot minimum="0" maximum="1.0" terminal="png"/>
<Column symbol-ref="cell.id" celltype="Angioblasts"/>
</HistogramLogger>
</Disabled>
-->
<DependencyGraph format="svg" exclude-plugins="Gnuplotter, CPM" exclude-symbols="cell, cell.center, str_2"/>
</Analysis>
</MorpheusModel>
Downloads
Files associated with this model: